Dataset to GDC Schema Alignment
Comprehensive guide for mapping biomedical datasets to the GDC schema standard
Installation 🛠️
For system-specific installation instructions:
- AMD64 architecture: Follow the Linux/Unix installation guide
- ARM64 architecture (Apple Silicon): Follow the MacOS installation guide
Introduction 📚
This guide demonstrates the process of aligning a biomedical dataset with the Genomic Data Commons (GDC) schema using BDIViz. The GDC maintains standardized data repositories that require specific formatting and schema compliance for successful data submission and integration.
Step 1: Dataset Preparation and Task Initialization 🚀
- Download the Clark et al. sample dataset
- Navigate to
http://localhost:3000in your web browser - Select “New Matching Task” from the navigation bar
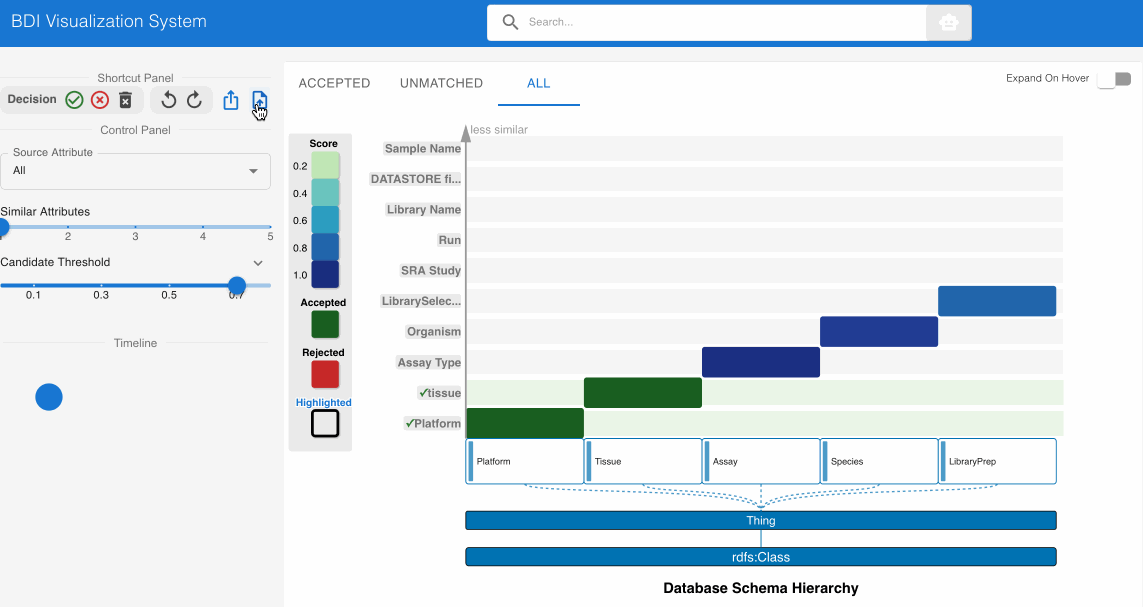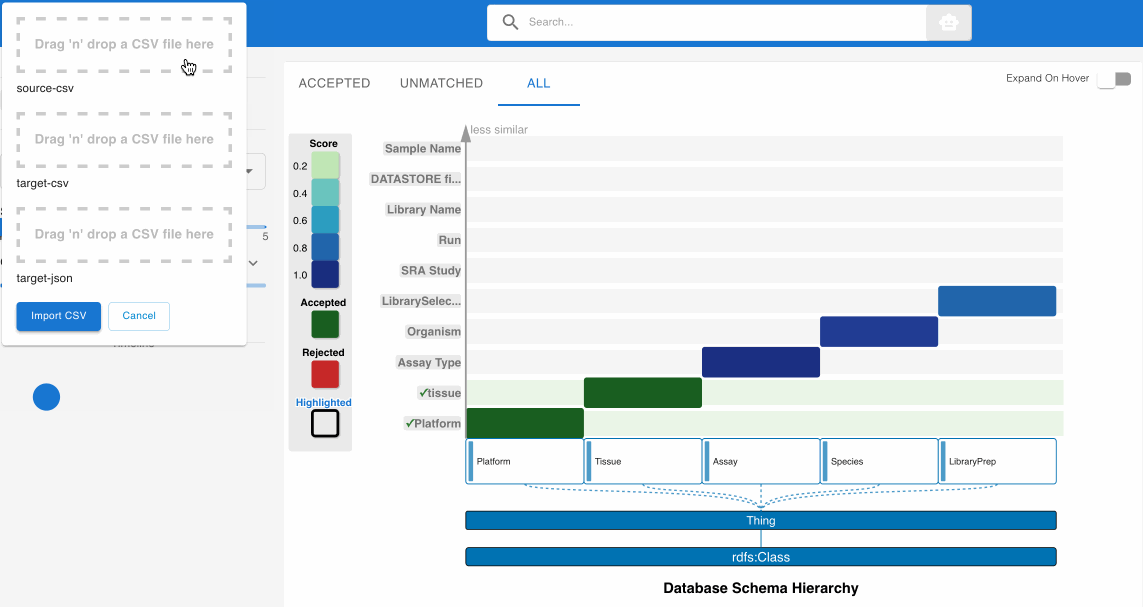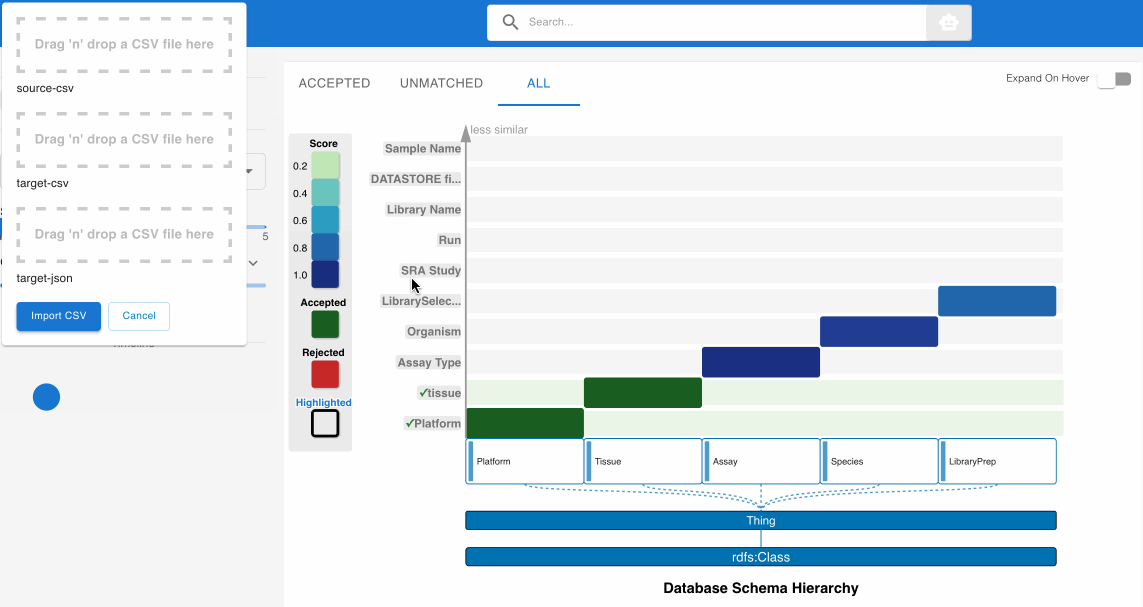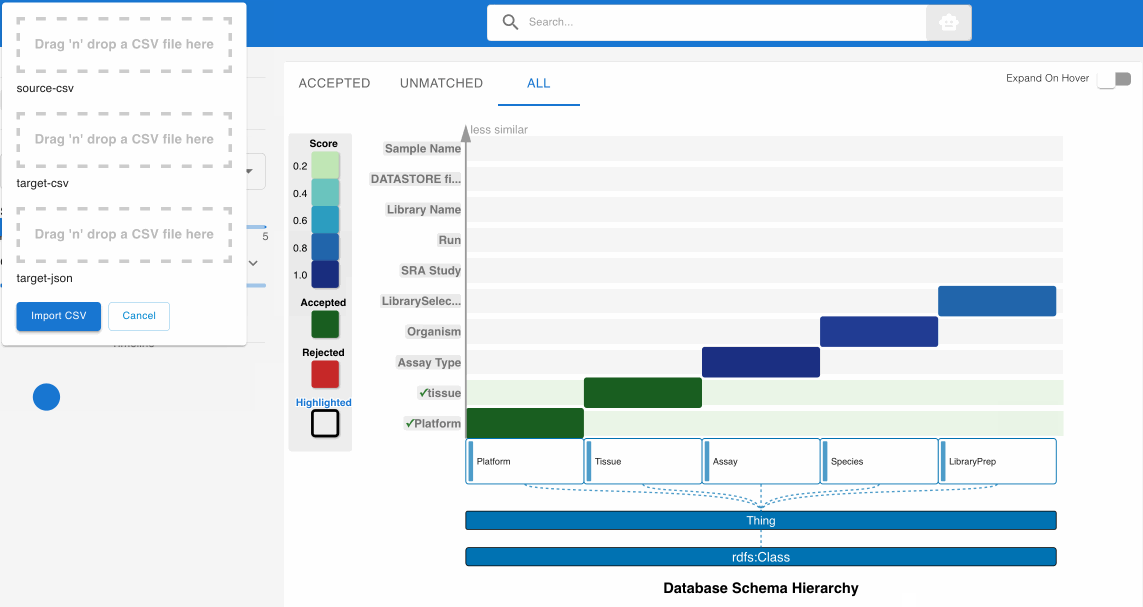
- Upload the Clark dataset CSV file to the Source CSV File drop zone
- Leave the Target CSV File and Target Schema JSON File fields empty to utilize the default GDC v3.3.0 schema
- Click “Import CSV” to initiate the matching process
Step 2: Validating High-Confidence Matches ✅
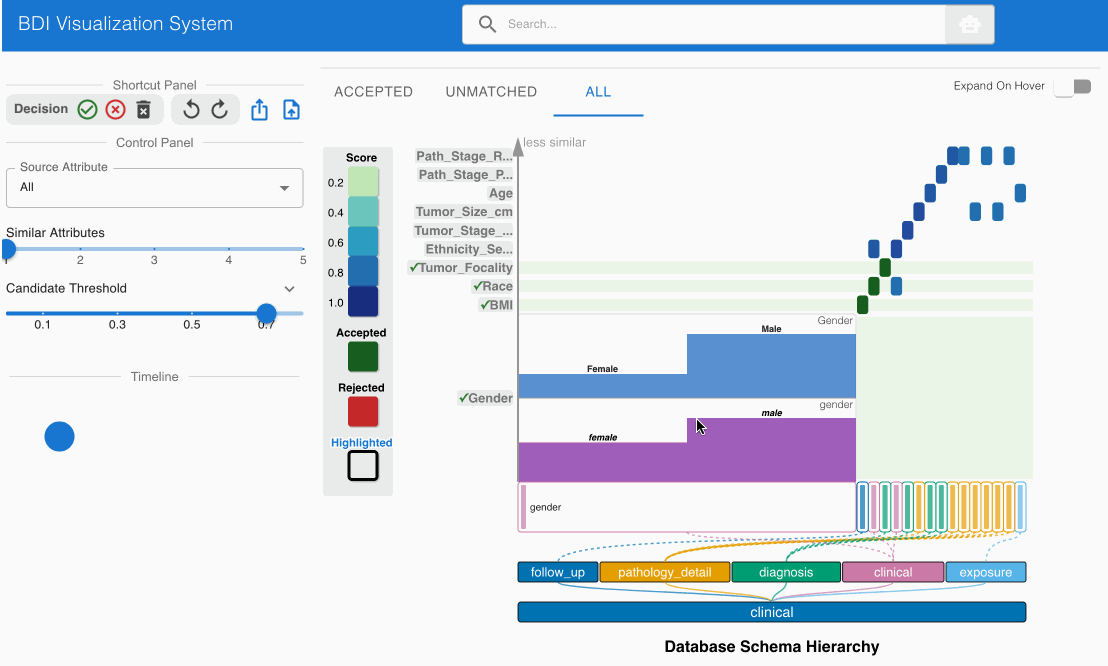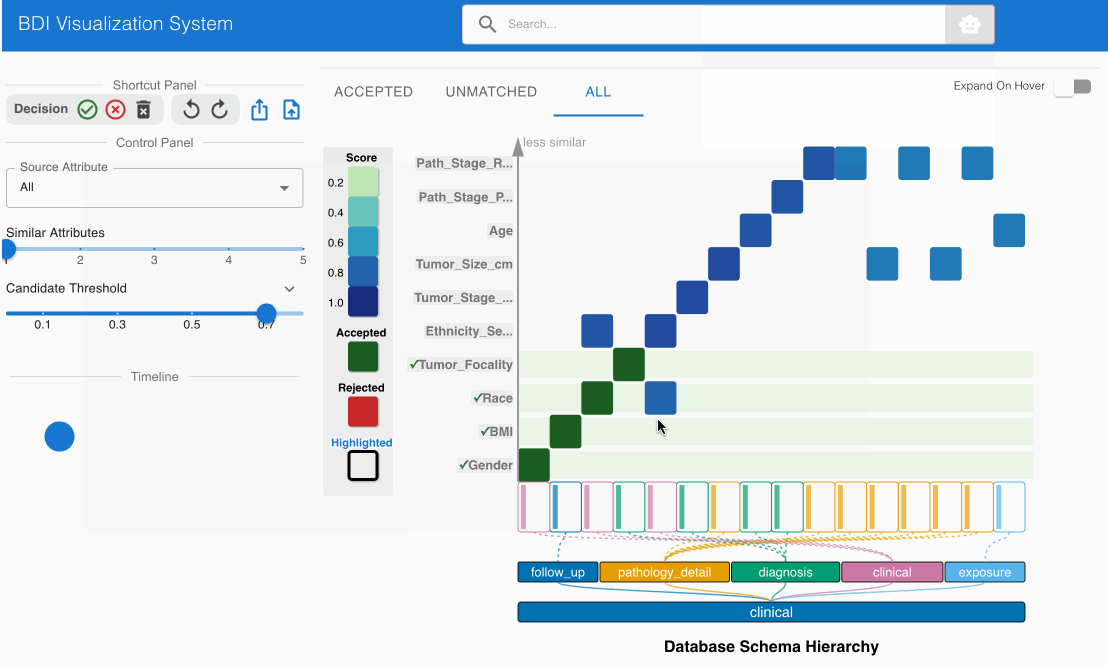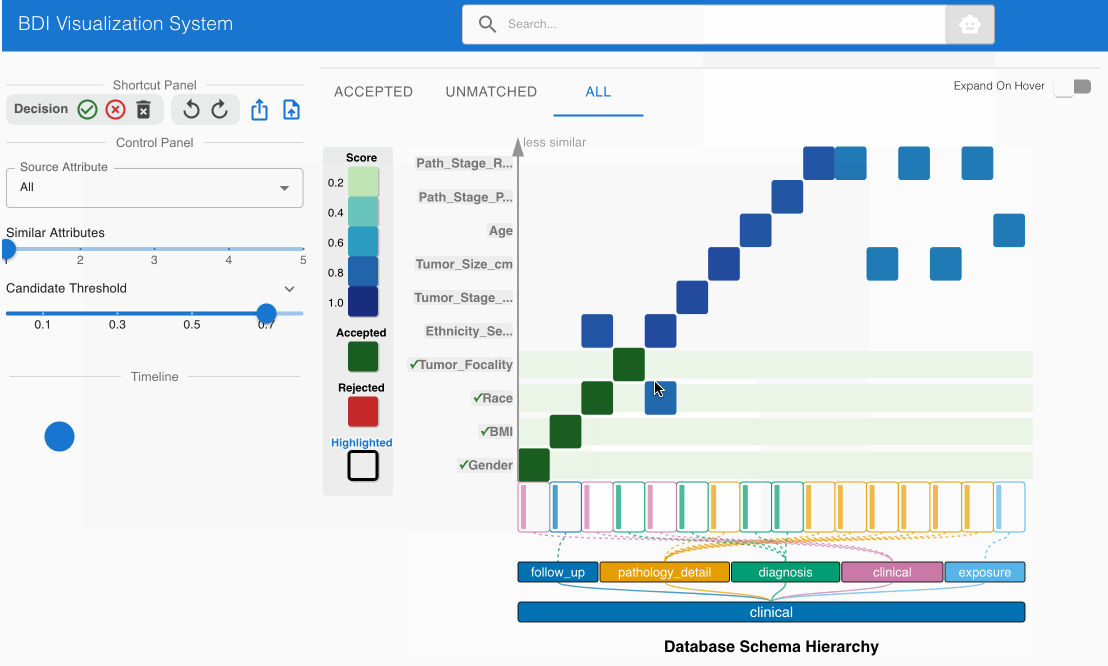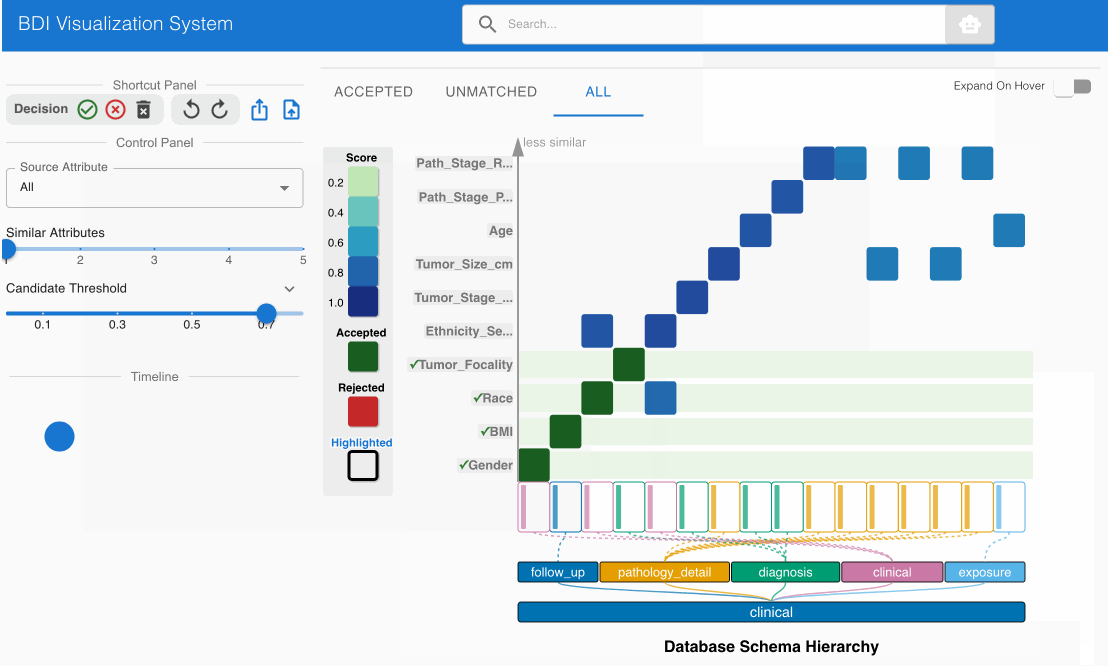
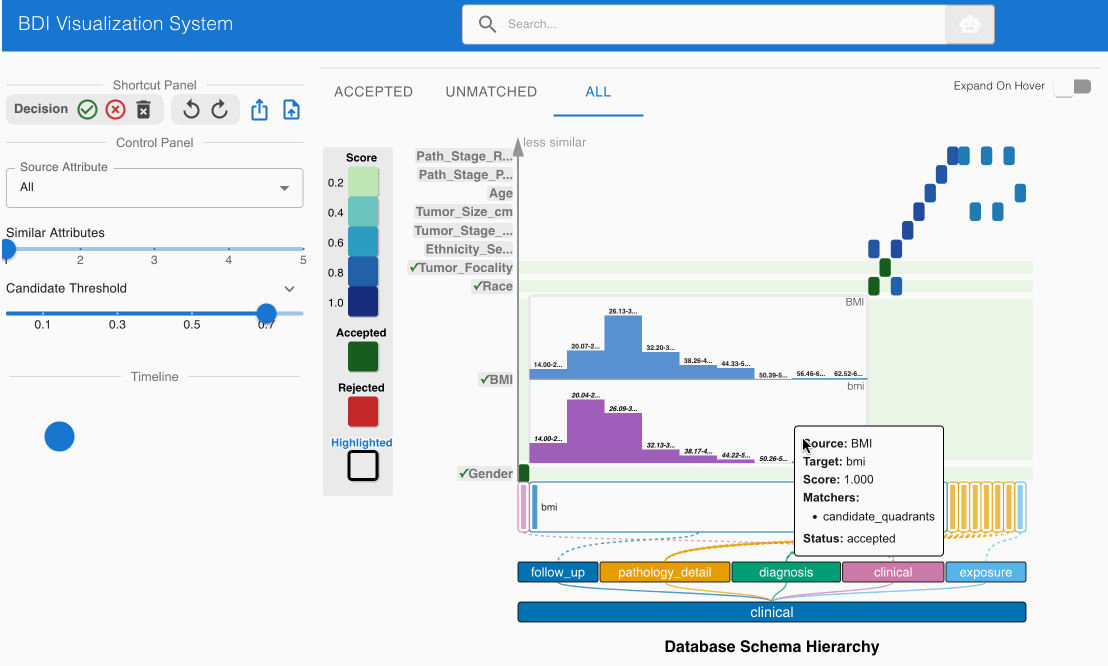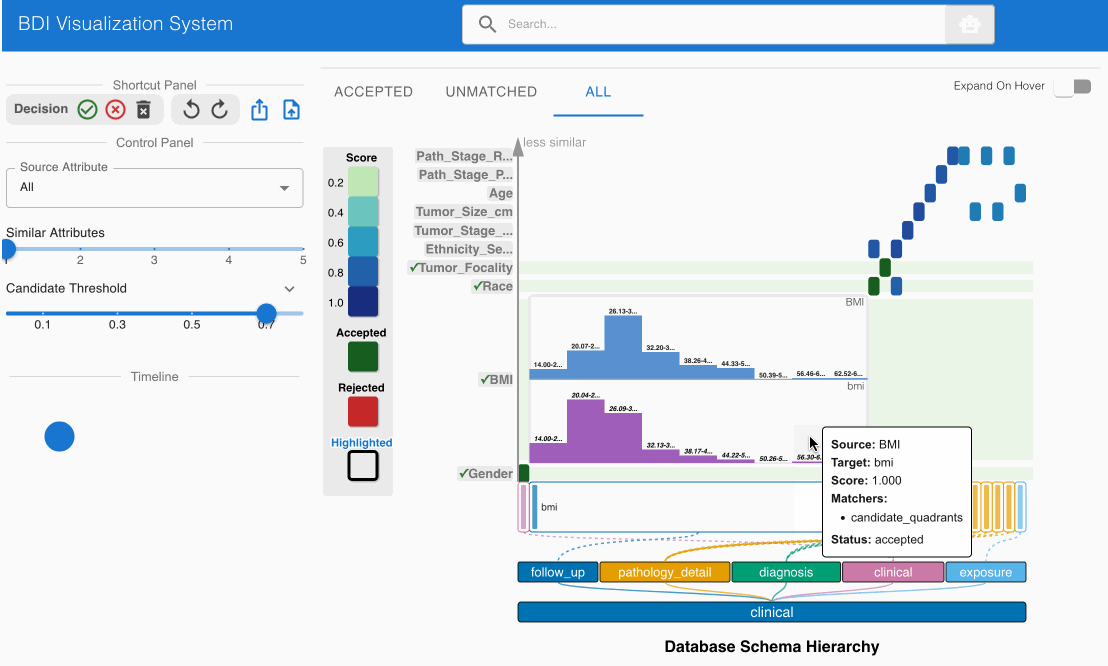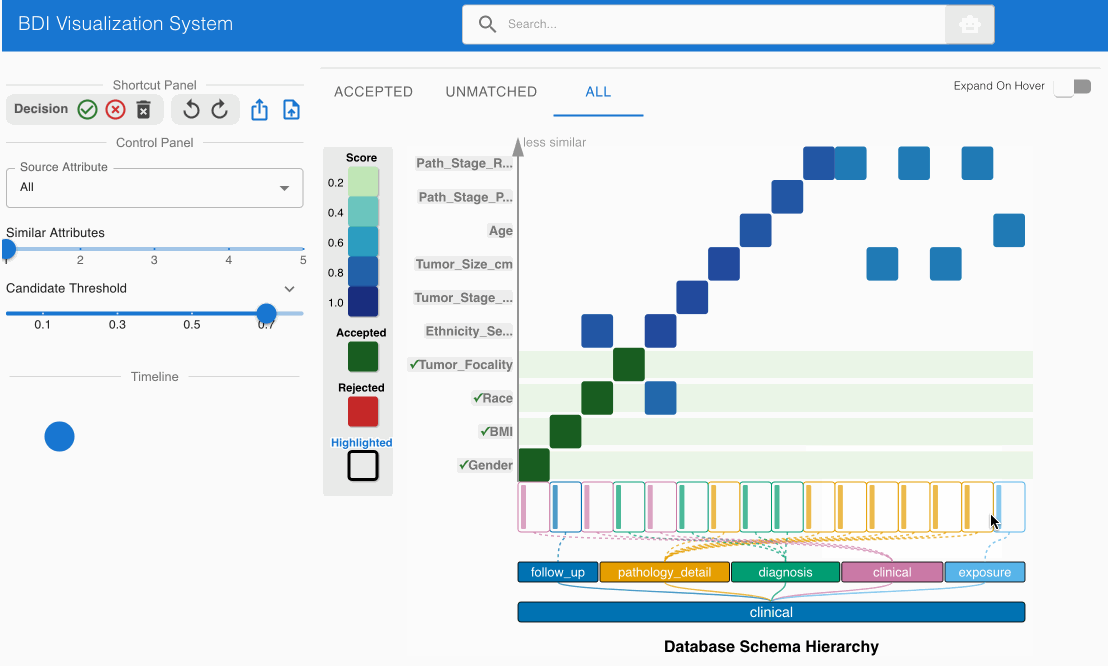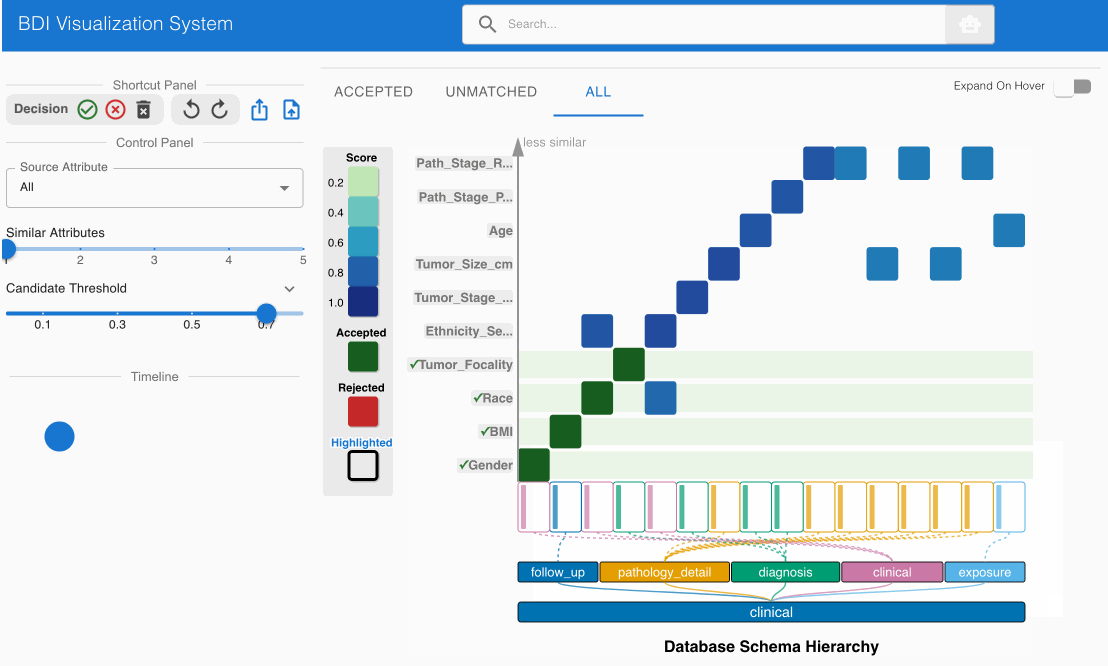
The system automatically identifies and suggests high-confidence matches, indicated by darker cells in the heatmap. Review these initial suggestions:
Direct Attribute Matches:
- Locate the matching candidate Gender (source) -> gender (GDC) in the heatmap cell
- Click the heatmap cell to expand the embedded node
- The embedded node contains unique value distributions for both attributes
- Verify that both attribute names and values align (Male → Male, Female → Female)
- Note that high-confidence matches are pre-accepted (indicated by green cells with checkmarks)
Numerical Attribute Alignment:
- Examine the matching candidate BMI (source) -> bmi (GDC) in the heatmap cell
- Click the heatmap cell to expand the embedded node
- The embedded node shows value bin distributions for numerical data
- Observe that both attribute names and numerical distributions correspond appropriately
- This match should also be pre-accepted (green cell with checkmark)
Step 3: Explore Complex Attribute Matches 🧩
Address more nuanced matching scenarios that require human judgment:
Evaluating Semantic Differences:
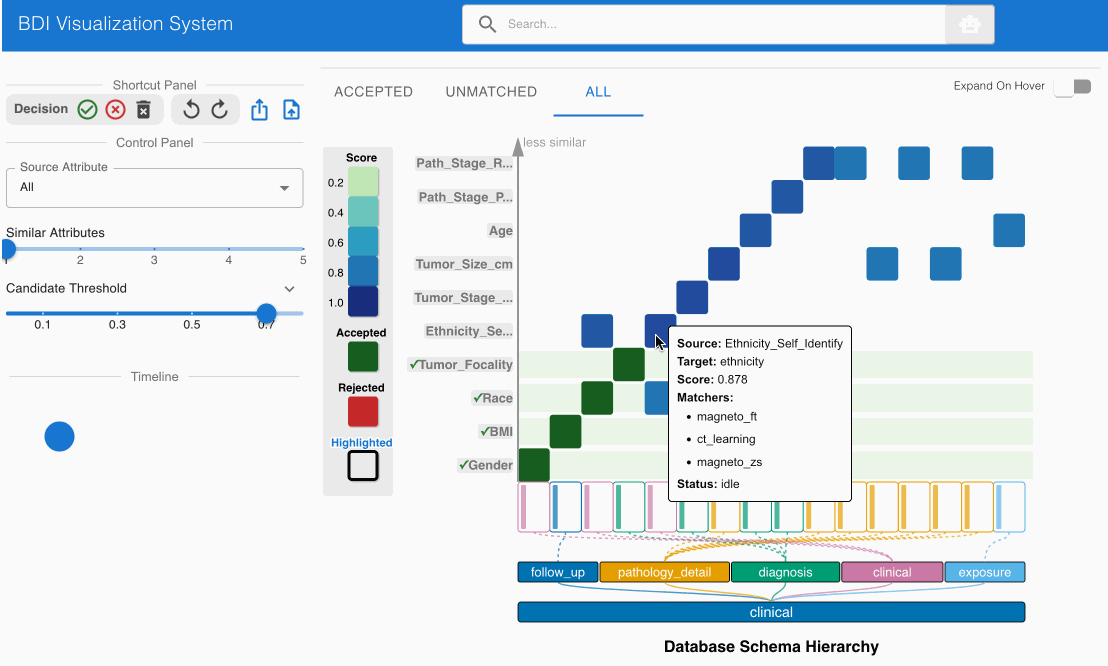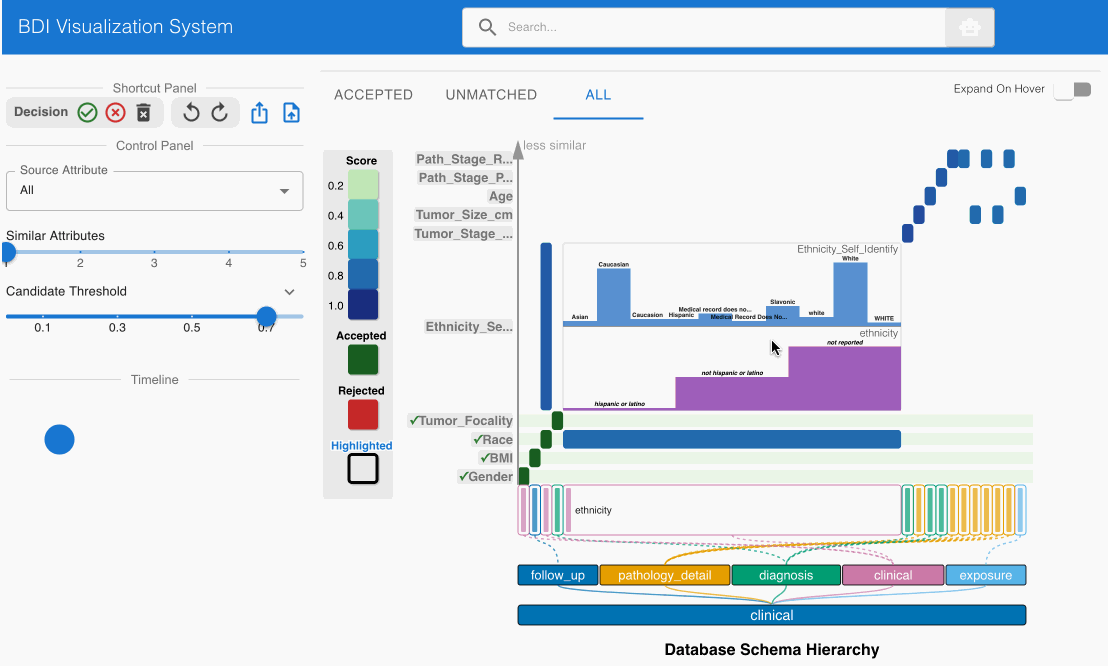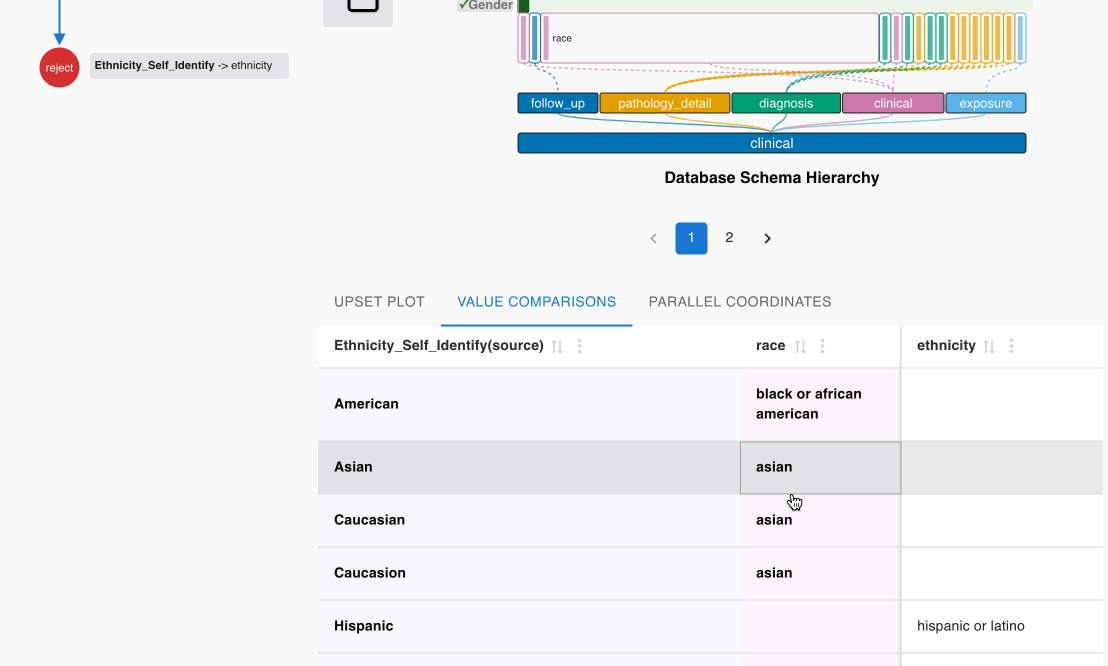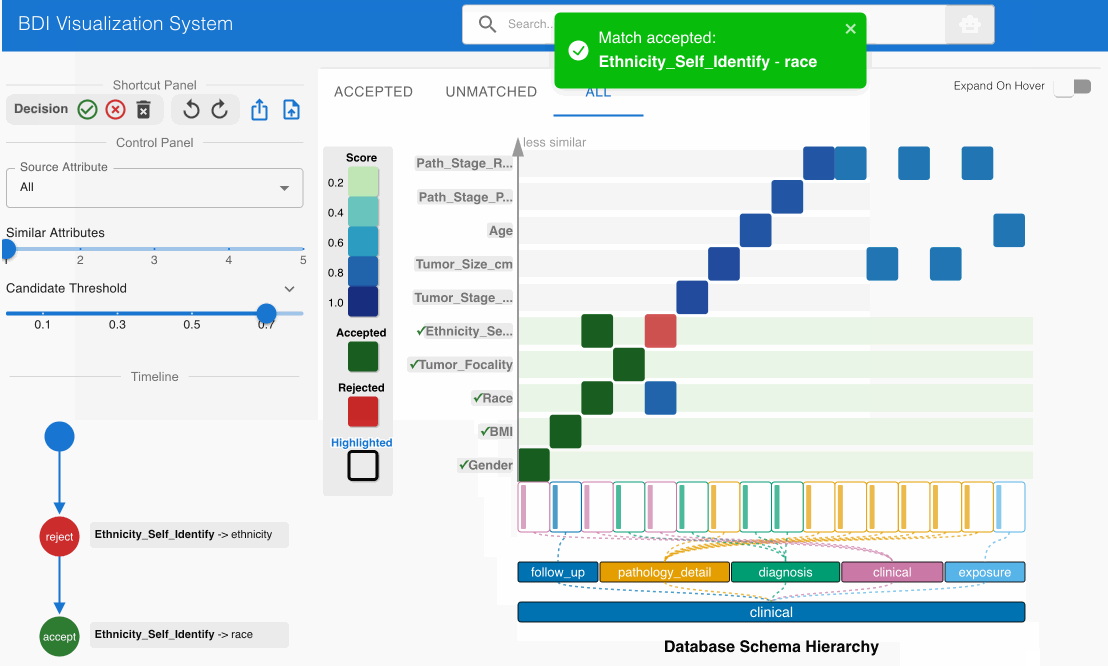
- Locate the matching candidate Ethnicity_Self_Identify (source) -> ethnicity (GDC) in the heatmap cell
- Click the heatmap cell to expand the embedded node
- Scroll down to check the value comparisons table in the embedded node
- Note the significant value discrepancies:
- Source values include “Asian”, “White”, etc.
- GDC ethnicity values include “hispanic or latino”, “not hispanic or latino”, etc.
- Click the Reject (✕) button on the top left Shortcut Panel to invalidate this incorrect match
Identifying Cross-Category Matches:
- Examine the matching candidate Ethnicity_Self_Identify (source) -> race (GDC) in the heatmap cell
- Click the heatmap cell to expand the embedded node
- Scroll down to check the value comparisons table in the embedded node
- Confirm that the values correspond appropriately:
- “Asian” in source aligns with “asian” in GDC
- “White” in source aligns with “white” in GDC
- Click the Accept (✓) button on the top left Shortcut Panel to confirm this cross-category match
Step 4: Leverage the LLM Agent for Complex Matching Decisions 🤖
The integrated LLM Agent provides valuable insights for challenging attribute matches:
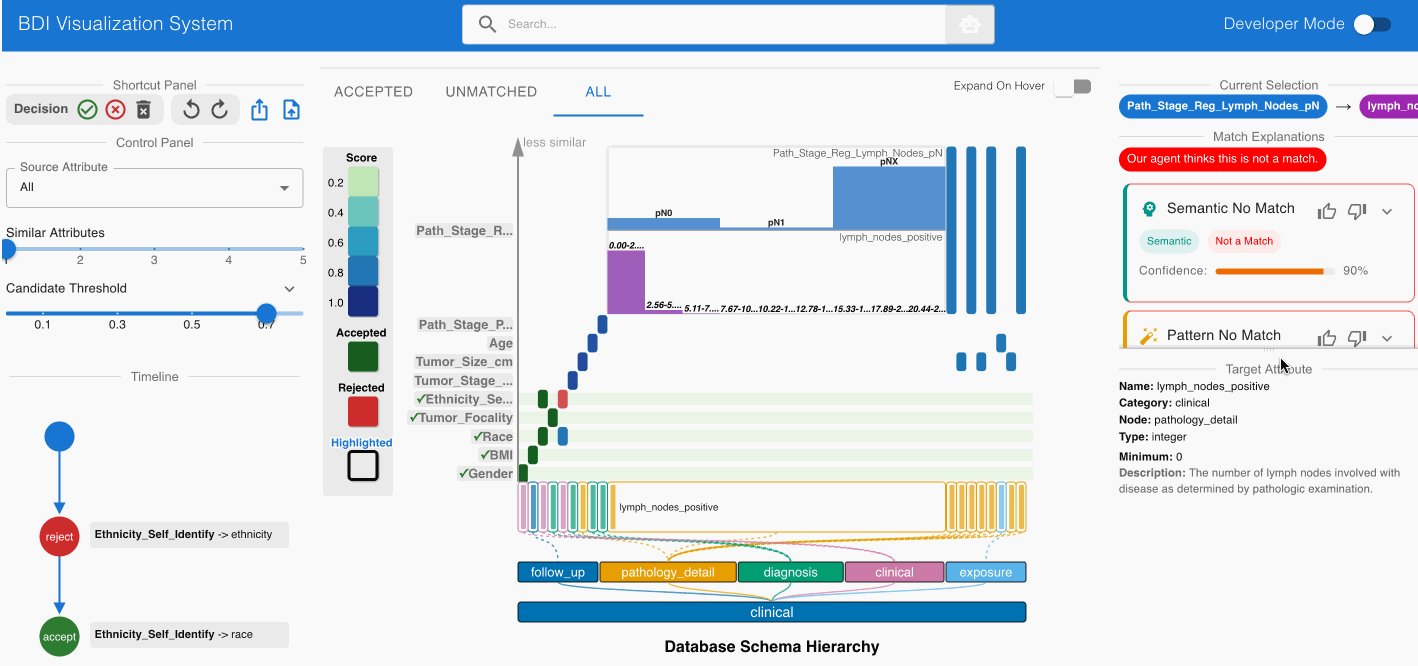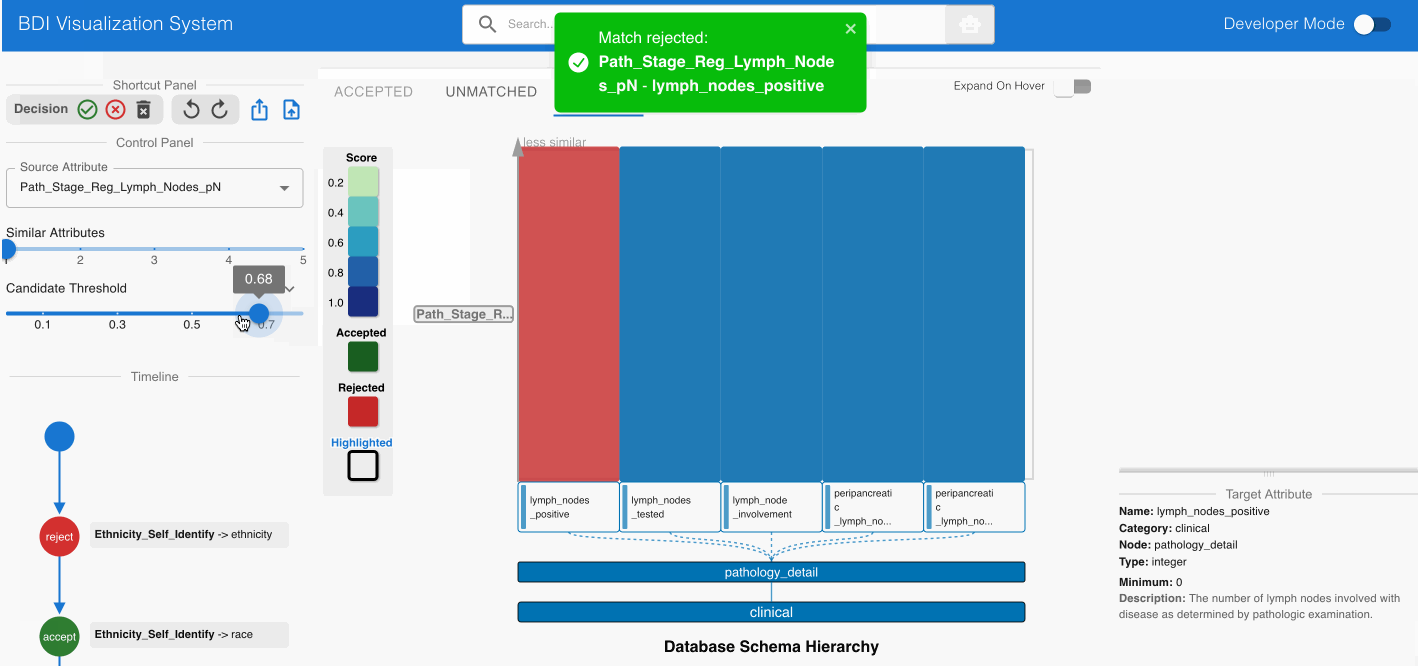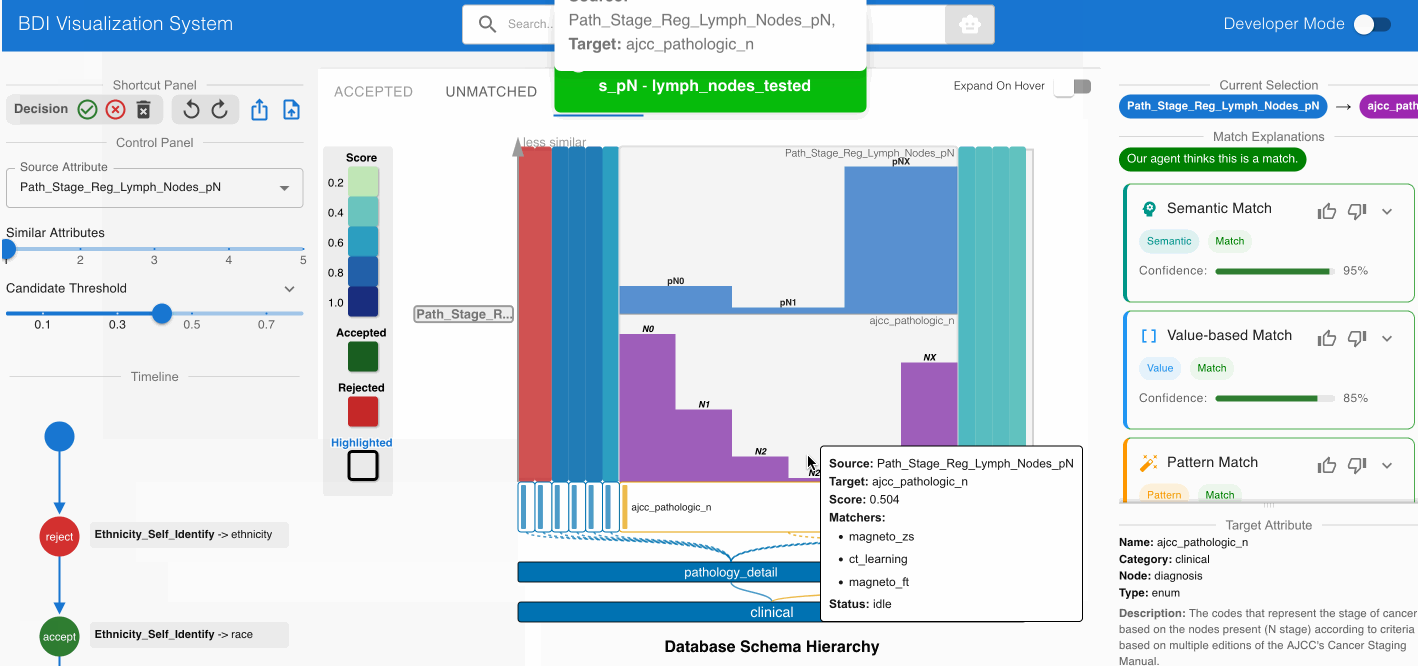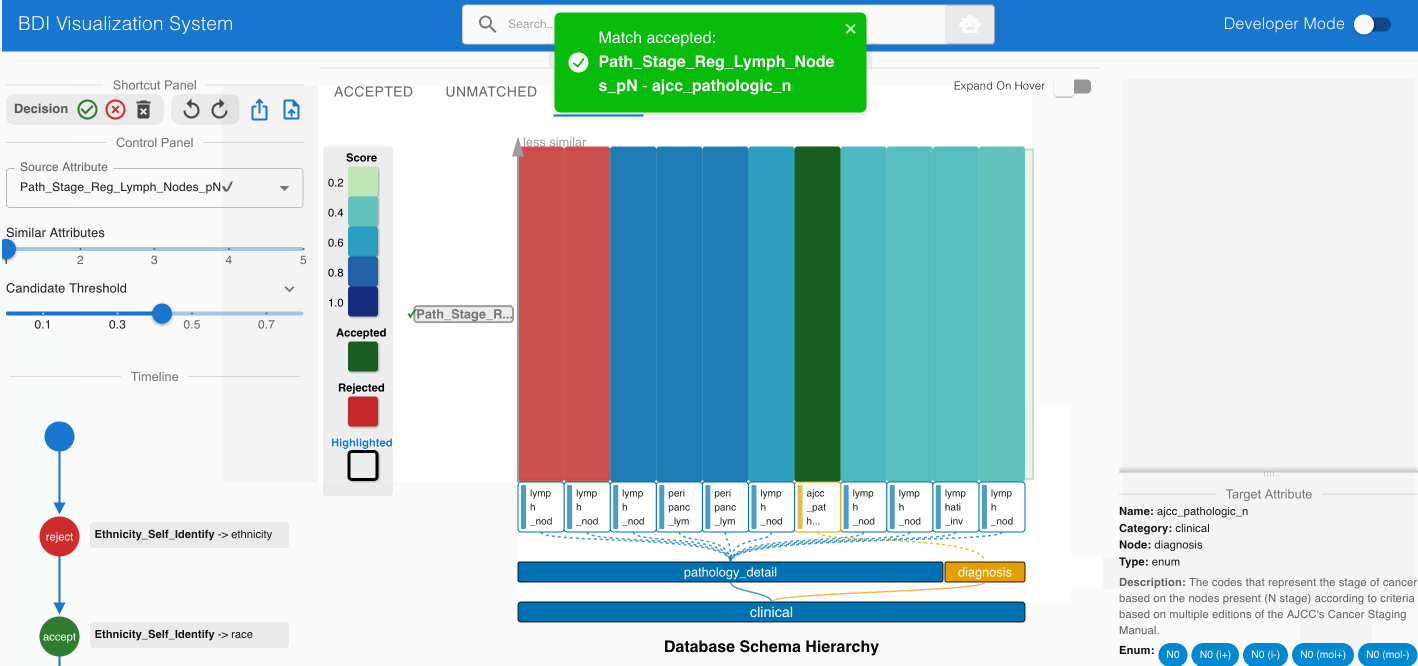
Evaluating Pathological Staging to Lymph Node Count:
- Examine the potential match Path_Stage_Reg_Lymph_Nodes_pN (source) -> lymph_nodes_positive (GDC)
- Consult the Agent Panel on the left side of the interface
- Review the “Semantic No Match” card which indicates:
- Source attribute represents a classification system (pN0, pN1, etc.)
- Target attribute represents a numerical count of positive lymph nodes
- Based on this analysis, reject this match using the Shortcut Panel
Distinguishing Staging from Testing Metrics:
- Evaluate the potential match Path_Stage_Reg_Lymph_Nodes_pN (source) -> lymph_nodes_tested (GDC)
- Expand the “Pattern No Match” card in the Agent Panel
- Note the critical insight that source values (pN0, pN1, pNX) represent staging categories
- Confirm these do not align with the expected numeric values for lymph nodes tested
- Reject this match using the Shortcut Panel
Identifying Non-Obvious Semantic Equivalence:
- Assess the potential match Path_Stage_Reg_Lymph_Nodes_pN (source) -> ajcc_pathologic_n (GDC)
- Despite different naming conventions, examine the value distributions
- Consult the “Semantic Match” card in the Agent Panel
- Verify that both attributes represent pathological lymph node staging in cancer systems
- Accept this match using the Shortcut Panel
Step 5: Systematic Review of All Source Attributes 🔍
Methodically evaluate each remaining source attribute:
- Prioritize heatmap cells with darker shading (indicating stronger match candidates)
- Click each heatmap cell to expand the embedded node and evaluate both attribute names and value distributions
- Utilize the top left Shortcut Panel to:
- Accept (✓) valid and appropriate matches
- Reject (✕) invalid or misaligned matches
- Discard (🗑️) attributes that should be excluded from the mapping process
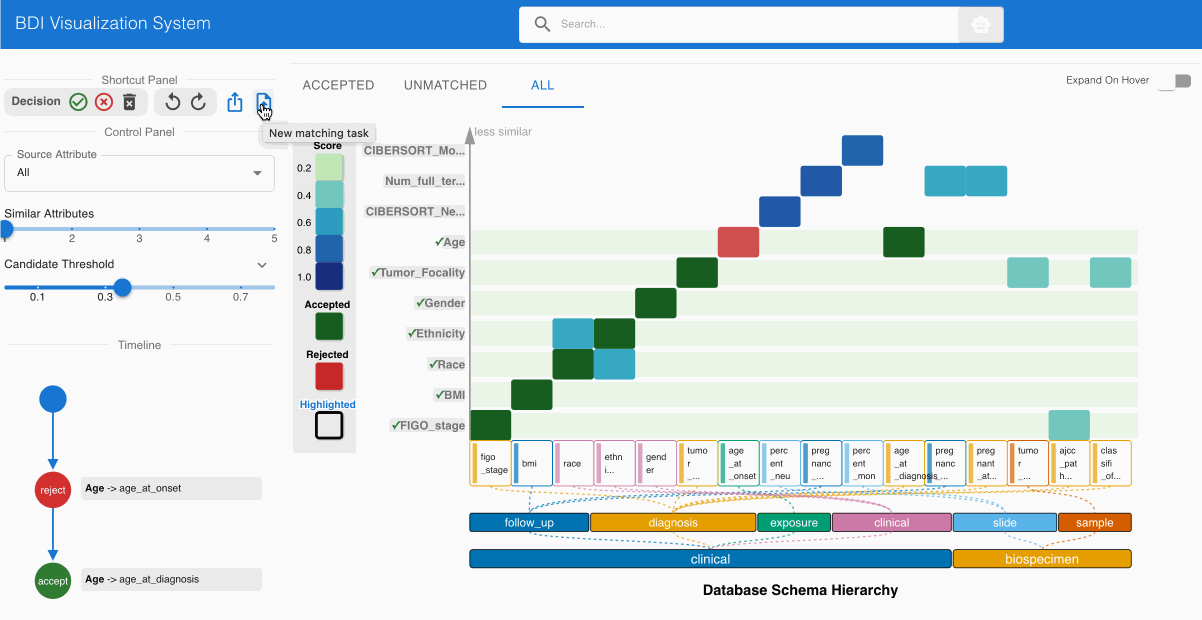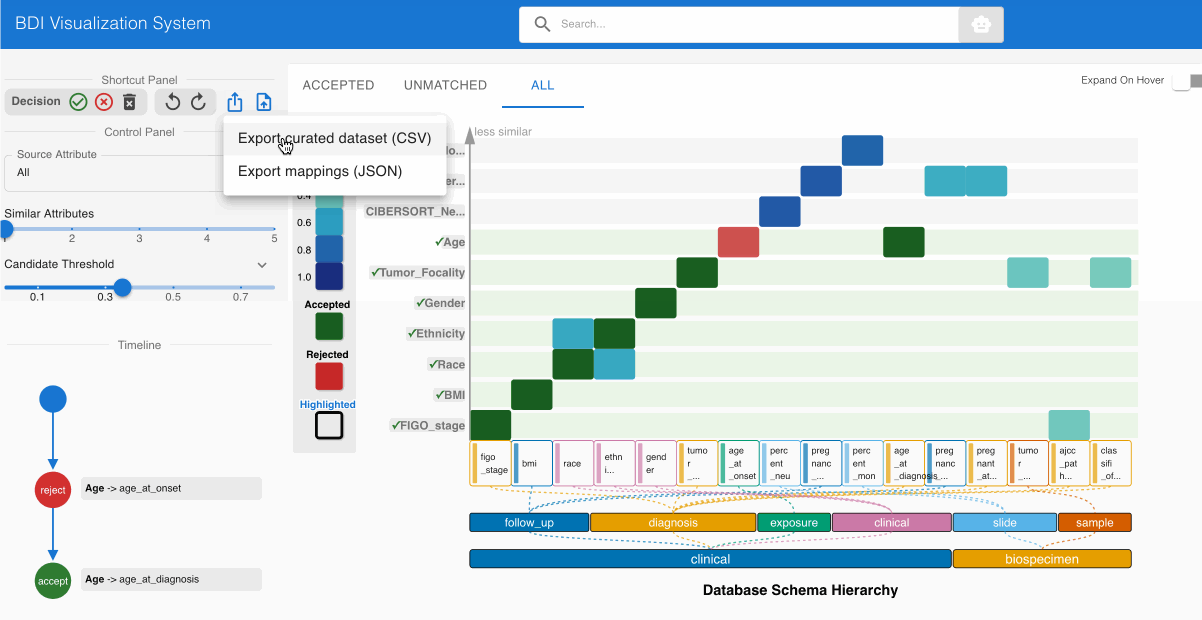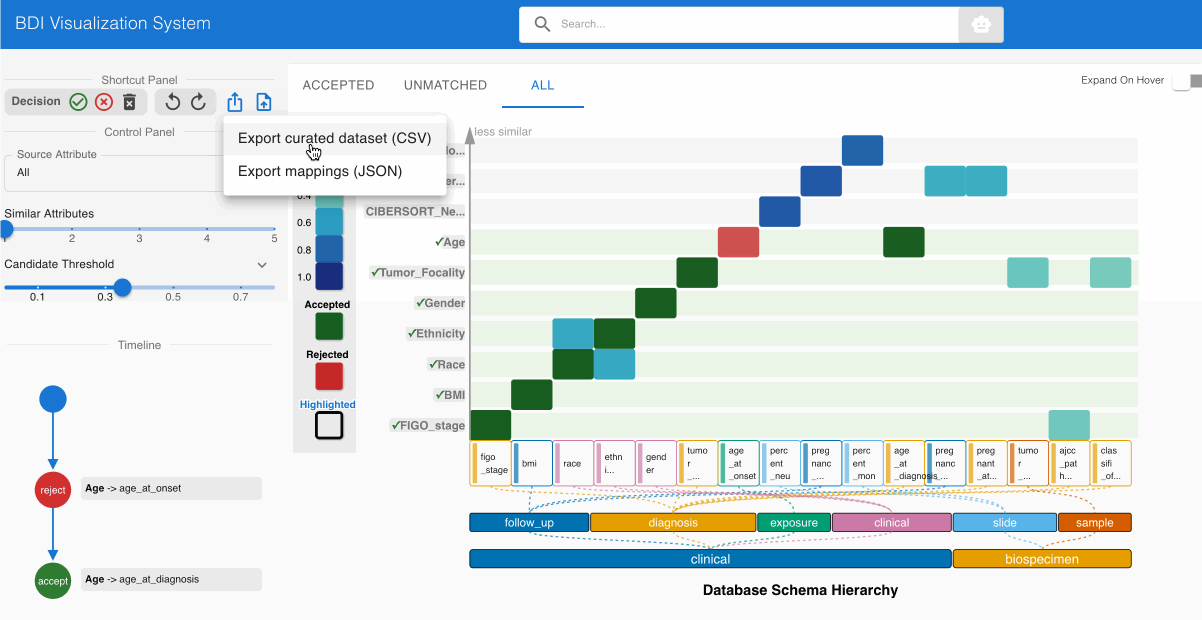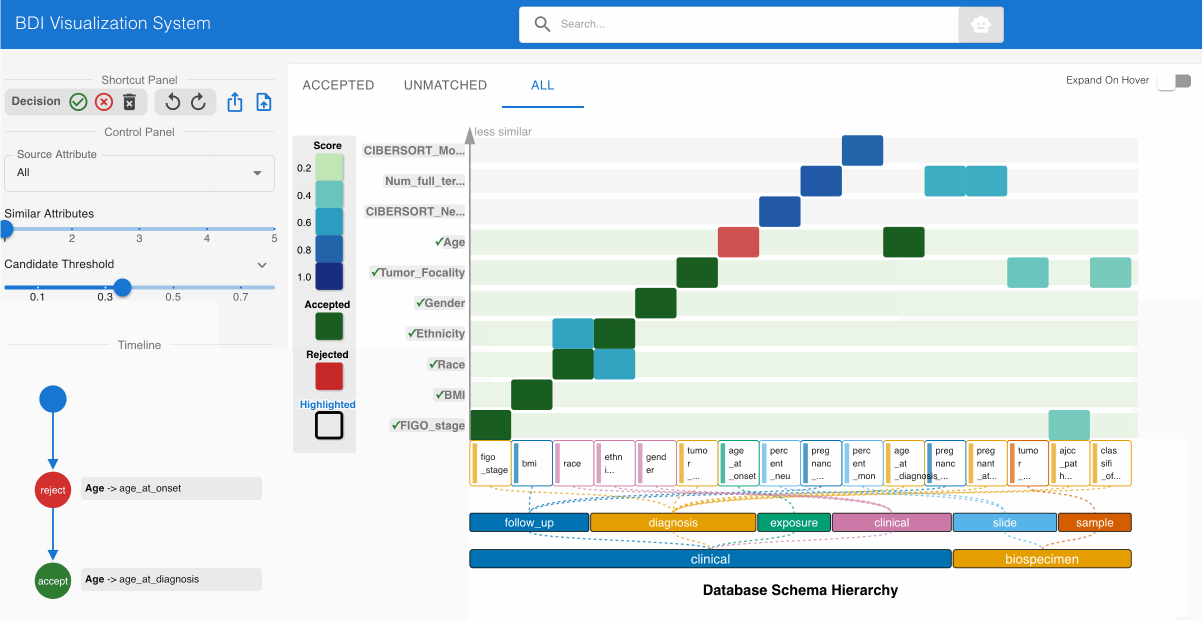
Step 6: Exporting the GDC-Compliant Dataset 📤
After completing the evaluation process:
- Select the Export button from the top left Shortcut Panel
- Choose CSV format to generate a dataset with GDC-compliant column names
- Save the transformed file to your local system
Further Resources 📖
For advanced functionality and detailed operational guidance, please refer to the comprehensive User Manual.